Thoroughly analyzing TS Inter 2nd Year Botany Model Papers and TS Inter 2nd Year Botany Question Paper March 2018 helps students identify their strengths and weaknesses.
TS Inter 2nd Year Botany Question Paper March 2018
Time : 3 Hours
Max. Marks: 60
Note: Read the following instructions carefully:
- Answer all questions of Section ‘A’. Answer any six questions out of eight in Section ‘B’ and answer any two questions out of three in Section ‘C.
- In Section ‘A’, questions from Sr. Nos. 1 to 10 are of ‘Very Short Answer Type”. Each question carries two marks. Every answer may be limited to 5 lines. Answer all the questions at one place in the same order.
- In Section ‘B’, questions from Sr. Nos. 11 to 18 are of “Short Answer Type”. Each question carries four marks. Every answer may be limited to 20 lines.
- In Section ‘C’, questions from Sr. Nos. 19 to 21 are of “Long Answer Type’. Each question carries eight marks. Every answer may be limited to 60 lines.
- Draw labelled diagrams wherever necessary for questions in Section ‘B’ and ‘C’.
Section – A
10 x 2 = 20
Note: Answer all questions. Each answer may be limited to 5 lines.
Question 1.
Define Hydroponics.
Answer:
The technique of growing plants in a specified nutrient solution in complete absence of soil is known as Hydroponics.
Question 2.
How does guttation differ from transpiration?
Answer:
| Guttation | transpiration |
| 1) The loss of water in the form of liquid or drops. | 1) The loss of water in the form of water vapour. |
| 2) It occurs usually at night. | 2) It occurs usually during the daytime. |
| 3) It occurs through vein endings. | 3) It occurs through tomato. |
| 4) It is an uncontrolled process. | 4) It is a controlled process. |
Question 3.
Define biotechnology?
Answer:
It is a science which utilises properties and uses of microorganisms or exploits cells and the cell constituents at the Industrial level for generating useful products essential to life and human welfare.
Question 4.
What are pleomorphic bacteria? Give an example.
Answer:
Some bacteria keep on changing their shape depending upon the type of nutrients available and the environment are called pleomorphic bacteria. Ex: Acetobacter.
Question 5.
Explain the terms, phenotype, and genotype.
Answer:
The physical or external appearance of a character is called Phenotype. The genetic makeup of an individual is called Genotype.
Question 6.
What are the components of a nucleotide?
Answer:
A nitrogenous base, a pentose sugar, and a phosphate group.
Question 7.
Why does ‘swiss cheese’ have big holes? Name the bacteria responsible for it.
Answer:
The big holes in ‘Swiss cheese’ are due to the production of large amounts of CO2. Propionibacterium sharmani is responsible for it.
![]()
Question 8.
What is the difference between exons and introns?
Answer:
| Exons | Introns |
| 1) Coding or expressed sequences. | 1) Intervening sequences. |
| 2) They do not appear in nature or processed RNA. | 2) They appear in nature or processed RNA. |
Question 9.
Name the nematode that infects the roots of tobacco plants. Name the strategy adopted to prevent this infestation.
Answer:
Meloidegyme incognita. A morel strategy adopted to prevent this infestation is process of RNA interference (RNA1).
Question 10.
Name a microbe used for statin production. How do statins lower blood cholesterol level?
Answer:
Monascus purpure us. Statins act by competitively inhibiting the enzyme responsible for the synthesis of cholesterol.
Section – B
6 x 4 = 24
Note: Answer any SIX questions. Each answer may be limited to 20 lines.
Question 11.
What is meant by plasmolysis? How is it practically useful to us?
Answer:
The phenomenon of shrinkage of protoplast due to osmotic diffusion of water from the cells into the surrounding environment is called plasmolysis. When a cell is placed in high concentrated solution, water moves out through osmosis. As a result, the protoplasm undergoes shrinkage leading to the separation of the plasma membrane from the cell wall in the corners called incipient plasmolysis. The salting of pickles and preserving of fish and meat in salt are good examples of practical applications.
Question 12.
Define RQ. Write a short note on RQ.
Answer:
The respiratory quotient is defined as the ratio between the volume of CO2 released to volume of 02 absorbed during respiration.
It is measured by Ganong’s respirometer.
Number of CO2 molecules released
RQ. = \(\frac{\text { Number of } \mathrm{CO}_2 \text { molecules released }}{\text { Number of } \mathrm{O}_2 \text { molecules absorbed }} \)
The RQ. value depends on the type of respiratory substrate used.
1. Carbohydrates: When carbohydrates re used as substrates, the volume of CO2 molecules released are equal
to the volume of O2 molecules consumed. So the RQ. value is one
C6H12O6 +6O2 + 6H2O → 6CO2 + 12H2O + energy
RQ = \(\frac{6 \mathrm{CO}_2}{6 \mathrm{O}_2}=\frac{6}{6}\) = 1
2. Fats: When fats are used in Respiration, the RQ. value is less than one because the volume of CO2 molecules
released are less than the amount of O2 molecules consumed.
2(C51H98O6) + 145O2 → 102CO2 + 98 H2O + energy
RQ = \(\frac{102}{145}\) = 0.7
3. Proteins: When proteins are used as respiratory substrates, RQ value would be about 0.9.
Question 13.
Write briefly about enzyme inhibitors.
Answer:
The chemicals that can shuts off enzyme activity are called enzyme inhibitors. They are of three types.
A. Competitive inhibitors: Substances which are closely resemble the substrate molecules and inhibits the activity of the enzyme are called Competitive inhibitors. Ex: Inhibition of Succinic dehydrogenase by Malonate which closely resembles the substrate Succinate.
B. Non-competitive inhibitors: The inhibitors which have no structural similarity with the substrate and bind to an enzyme at locations other than the active sites, so that the globular structure of the enzyme is changed are called Non-competitive inhibitors. Ex: Metal ions of Copper, Mercury.
C. Feedbck inhibitors: The end product of a chain of enzyme catalysed reactions inhibits the enzyme of the first reaction as part of homeostatic control of metabolism is called feedback inhibitors. Ex: During respiration, accumulation of Glucose – 6phosphate occurs, inhibits the Hexokinase.
![]()
Question 14.
Write a note on agricultural/horticultural applications of Auxins.
Answer:
- IBA, NAA, and IAA help to initiate rooting in stem cuttings. widely used for plant propagation in horticulture.
- Auxins like 2, 4- D, 2, 4, 5 – T act as herbicides and kills broad-leaved dicot weeds to prepare weed-free lawns.
- Auxins stimulate fruit growth. Ex: Tomato.
- Auxins induce flowering in pineapple.
- Auxins prevent premature leaf and fruit drop.
Question 15.
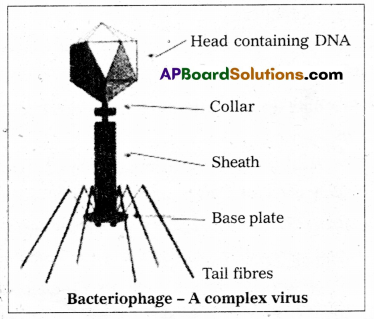
Explain the structure of T-even bacteriophages.
Answer:
- The viruses which attack bacteria are called Bacteriophages. They were discovered by Tvort (1915).
- Felix ‘d’Herelle (1917) coined the term Bacteriophage.
- Bacteriophages are tadpole-shaped with a large head and a tail.
- The head is hexagonal and is capped by hexagonal pyramid. measures about 65 x 95 nm.
- The head is formed with several cap-someres. each of which is a single protein. The head protein forms a semipermeable membrane enclosing the folded double-stranded DNA which is 1000 times longer than the phage.
- The tail is composed of several parts present around central core.
Question 16.
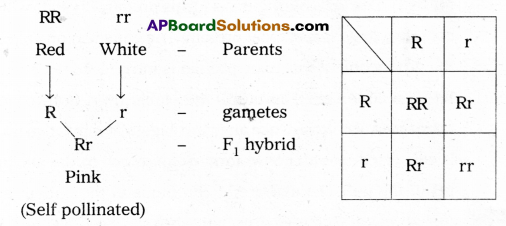
Explain incomplete dominance with a example.
Answer:
Incomplete dominance: It is the condition where one allele of a gene is not completely dominant over the other allele and results in the heterozygotes having phenotype different from the dominant and recessive homozygotes.
Ex: In a cross between true-breeding read flowered plant (RR) and true breeding white-flowered plant (rr), the F1 was
pink (Rr). When the R1 was self-pollinated, the F2 resulted in the ratio of
RR: Rr: rr
1: 2: 1
Red Pink White
Here the genotype ratios were as in monohybrid cross of Mendel but the phenotypic ratio had changed from 3: 1 because R was not completely dominant over ‘r’ and is possible to distinguish ‘Rr’ as pink from ‘RR and ‘rr.
Question 17.
Write the important features of genetic code.
Answer:
- The codon is triplet, of 64 codons, 61 codons code for 20 amino acids and 3 codons do not code for any amino acids (stop codons, UAA, UAG, UGA.)
- One codon codes for only one amino acid, hence it is unambiguous and specific. .
- Some amino acids are coded by more than one codon, hence the code is degenerate.
- The codon is read in mRNA in a contiguous fashion. There are no punctuations.
- The code is nearly universal. For Ex: UUU code for phenylalanine (phe) in bacteria and humans. .
- AUG codes for methionine and also acts as an initiator codon. (Dual role).
Question 18.
Give a brief account on Bt. cotton?
Answer:
Some strains of Bacillus thuringiensis produce proteins that kill certain insects such as lepidopterans (tobacco budworm, armyworm), coleopterans (beetles) and dipterans (files, mosquitoes). Bacillus thuringiensis forms protein Crystals which contain a toxic insecticidal protein. The gene responsible for the production of this toxic protein is introduced genetically into the cotton seeds protects the plants from Bollworm, a major pest of cotton. Once an insect Ingests the inactive toxin is converted into an active form of toxin due to the alkaline pH of the gut. The activated toxin binds to the midgut epithelial cells and creates pores that cause cell swelling and lysis and cause the death of the insect. Use of Bt. Cotton has led to 3-27% increase in cotton yield in countries where it is: grown. The toxin is coded by a gene named ‘cry’. The proteins encoded by the genes cry lAc and cry lAb control the cotton boll worms and cry to lAb controls corn borer.
Section – C
2 x 8 = 16
Note: Answer any two questions. Each answer may be limited to 60 lines.
Question 19.
Explain Calvin Cycle.
Answer:
Melvin Calvin, Andrew Benson, and James Basham discovered this pathway in chlorella and hence named as Calvin cycle. It is also called the Photosynthetic Carbon Reduction cycle [PCR cycle] or reductive pentose phosphate pathway.
The Calvin cycle can be described in three stages:
- Carboxylation
- Reduction and
- Regeneration.
1) Carboxylation: In this, six CO molecules reacts with six molecules of RUBP to form 12 molecules of PGA in the presence of water and RUBISCO, an enzyme.
6CO2 + 6RUBP + 6H2O → 12 PGA
2) Reduction
1) 12 molecules of PGA are phosphorylated to 12 molecules of DPGA in the presence of phosphogly cerokinase.
12PGA+ 12ATP → 12DPGA+12ADP
2) 12 DPGA molecules are reduce to 12 G3P molecules in the presence of G3P dehydrogenase.
12 DPGA + 12 NADPH + H+ + 12G3P + 12NADP + 12H2PO4–
3) Regeneration: Of 12 G3P molecules, 2G3P molecules are transported from chloroplast to cytosol. Here, they are utilised in the synthesis of hexose and then sucrose. Remaining 10 G3P are utilised to regenerate RUBP molecules.
4) Of 10 G3P molecules, 4 are isomerised to 4 DHAP (Dihydroxy acetone phosphate) in the presence of Isomerase.
4G3P → 4DHAP
5) 2 molecules of G3P condense with 2 DHAP molecules to form fructose 1, 6 Biphosphate in the presence of aldolase.
2G3P + 2DHAP → 2F1, 6BiP+
6) 2 molecules of F1, 6Bip undergoes dephosphorylation to 1orm F6P in the presence of phosphotase.
2F1 6Bip → 2F6P + 2iP
7) 2 molecules of F6P combines with 2 molecules of G3P to form two molecules of Erythrose 4 phosphate and 2 molecules of xylulose – 5 – phosphate in the presence of transketolase.
2F6P + 2G3P → 2XMP + 2EMP
8) Two molecules of EMP combines with 2DHAP to form 2 molecules of SHDP (Sedoheptulose — 1, 7 biphosphate) in the presence of aldolase.
2 EMP+ 2 DHAP → 2 SHDP
9) Two SHDP molecules undergoes dephosphorylation to 2 SHMP in the presence of phosphatase inorganic phosphate is released.
2 SHDP → 2 SHMP = 2iP
10) 2SHMP molecules combines with 2G3P to form 2 molecules of ribose monophosphate (RMP) and 2 molecules of XMP in the presence of transketolase.
2SHMP + 2G3P → 2RMP + 2XMP
11) 4 molecules of XMP (7, 10) are epimerised to form 4 molecules of RUMP in the presence of epimerase.
4XMP → 4RUMP
12) 2 RMP molecules are isomerised to 2 RUMP molecules in the presence of Isomerase.
2RMP → 2RUMP
13) 6 RUMP molecules are phosphorylated to form 6 RUBP molecules in the presence of kinase.
6 RUMP + 6 ATP → 6 RUBP + 6 ADP
The overall equation is
6CO2 + 6RUBP + 6H2O + 18 ATP + 12 NADPH + H+ → Glucose + 6 RUBP + 18 ADP + 12 NADP+
Hence for every CO2 molecules entering into the Calvin, cycle, 3 ATP and 2 NADPH are required.
![]()
Question 20.
Explain briefly the various processes of recombinant DNA technology.
Answer:
Key tools are:
1. Restriction enzymes: Two enzymes reponsible for restricting the growth of Bacteriophage in Escherichia. coli were isolated In-the year 1963. One of these added methyl group to DNA and the Other The latter was called restriction endonuclease. The first restriction endonuclease. Hind II which Six DNA molecules at a particular pair by recognising a specific sequence of Six base pairs, called recognition sequence for Hind II. Today more than 900 restriction enzymes were isolated from over 200 strains of Bacteria each of which recognises a different recognition sequence.
E.CORI is a restriction enzyme in which, the first letter comes from the genus (escherichia) and the second two letters from the species of the prokaryotic cell (coil) the latter:‘R’ is derived from the name of strain. Roman numbers indicate the order in which the enzymes were isolated from the strain of Bacteria Restriction enzyme belong to a larger class of enzymes called nucleases.
They are of two types:
a) Exonucleases which remove nucleotides from the ends of the DNA.
b) Endonucleases which makes cuts at specific location within the DNA.
Most restriction enzymes cut the two stands of DNA double heix at different locations such a clevage is known as staggered cut. E.CORI recognises 5’ GAATT3 sites on the DNA and cut it between G & A results in the formation of sticky ends or cohesive end pieces. This stickyness of the ends facilitates the action of enzyme DNA ligase.
Cloning vectors: The DNA used as a camera for transferring a fragment of fòreign DNA into a suitable host is called vector. Vectors used for multiplying the foreign DNA sequences are called cloning vectors. Commonly used cloning vectors are plasmids, bacteriophages, cosmids, plasmids are extrachromosome circular DNA molecules present in almost all bacteria species. They are inheritable and carry few genes are easy to isolate and reintroduce into the bacterium (host).
Features required to facilitates cloning into a vector:
a) Origin of replication: (ori) This is a sequence from where replication starts and any piece of DNA when Linked to this sequence can be made to repliçate within host cells. It is also responsible for controlling the copy number of the linked DNA.
b) Selectable marker: In addition to tori the vector requires a selectable maker, which help in identifying and eliminating non-transformants and selectively permitting the growth of the any transformants normally, the genes encoding resistance to Antibiotics such as Ampicillin. Chloramphenicol, tetracycline or Kanamycin, etc., are useful selectable makers for E.coli.
c) Cloning sites: In order to link the alien DNA, the vector needs to have very few preferably single recognition sites for the restriction enzyme.
d) Molecular weight: The cloning vector should have low molecular weight.
e) Vectors for cloning genes in plants and animals: The tumour-inducing (Ti) plasmid of Agrobacterium tunifaciens has now been modified into a cloning vector such that it is no more pathogenic to plants. Similarly, retroviruses have also been disarmed and are now used to deliver desirable genes into animal cells.
Question 21.
You are a Botanist, working in the area of plant breeding. Describe the various steps that you will undertake to release a new variety.
Answer:
The main steps in breeding a new genetic variety of a crop are:
1) Collection of Variability: Genetic variability is the root of any breeding programme. Collection and preservation of all the different wild varieties, species, and relatives of cultivated species is a prerequisite for effective exploitation of natural genes available in the populations. The entire collection having all the diverse alleles for all genes in a given crop is called Germplasm collection.
2) Evaluation and selection of parents: The germplasm is evaluated so as to identify plants with desirable characteristics. The selected plants are multiplied and are used. Purelines are created wherever desirable.
3) Cross Hybridisation among the selected parents: After emasculation (Removal of Anthers from bisexual flower of a female -parent) the female flowers are enclosed in a polythene bag to prevent undesired cross-pollination. Pollen grains are collected from the male parent with the help of a brush and are transferred to the surface of the stigma and thus cross pollination is affected artificially.
4) Selection and Testing of superior recombinants: It in volves selecting among the progeny of hybrids, those plants that have the desired character combination. The selection process requires careful scientific evaluation of the progeny. Due to this, plants that the superior to both the parents are obtained. These are self-pollinated for several generations till they reach a homozygosity.
5) Testing, release, and commercialisation of new characters:
Characters: The newly selected lines are evaluated for their yield and other traits of quality, disease resistance, etc. It is done by growing these in research fields and recording their performance under ideal fertilizer application, irrigation, and other crop management practices. It is followed by testing the materials in farmers’ fields for at least 3 growing seasons at several places in the country, in all agroclimatic zones. Finally, they are distributed to farmers as a new variety.