Thoroughly analyzing TS Inter 2nd Year Botany Model Papers and TS Inter 2nd Year Botany Question Paper March 2019 helps students identify their strengths and weaknesses.
TS Inter 2nd Year Botany Question Paper March 2019
Time: 3 Hours
Max. Marks: 60
Note: Read the following instructions carefully.
- Answer All the questions of Section – A. Answer any six questions out of eight in Section – B and answer any two questions out of three in Section – C.
- In Section – A, questions from Sr. Nos. 1 to 10 are of ‘Very Short Answer Type”. Each question carries two marks. Every answer may be limited to 5 lines. Answer all the questions at one place in the same order.
- In Section – B, questions from Sr. Nos. 11 to 18 are of ‘Short Answer Type”. Each question carries four marks. Every answer may be limited to 20 lines.
- In Section – C, questions from Sr. Nos. 19 to 21 are of” Long Answer Type”. Each question carries eight marks. Every answer may be limited to 60 lines.
- Draw labelled diagrams, wherever necessary for questions in Section – B and C.
Section – A
10 x 2 = 20
Note: Answer all the questions. Each answer may be limited to 5 lines.
Question 1.
Name two amino acids in which sulphur is present.
Answer:
Cysteine, Cystine and Methionine.
Question 2.
With reference to transportation of food within a plant, what are source and sink?
Answer:
With reference to transportation of food, source is photos the sizing organs i.e., leaves and sink refers to consuming organs i.e., roots.
Question 3.
What is a plasmid? What is its significance?
Answer:
Plasmid is a small self-replicating, circular, double-stranded DNA molecule present in a bacteria in addition to main genetic material. They are used as agents (vectors) in modern Genetic engineering techniques.
![]()
Question 4.
What is point mutation? Give an example.
Answer:
Change in single base pair in the gene beta globulin chain results in the change of amino acid glutamate to valine. Ex : Sickle cell anaemia.
Question 5.
Who proved that DNA is genetic material? What is the organism they worked on?
Answer:
Alfred Hershey and Martha Chase (1952). They worked on the Bacterlo phases.
Question 6.
Define stop codon. Write the codons.
Answer:
The genetic codons which do not code for any amino acids is called stop codons. They are UAA, UAG and UGA.
Question 7.
Name any two artificially restricted plasmids.
Answer:
pBR 322 pUC 19, 101.
Question 8.
Can a disease be detected before its symptoms appear? Explain the principle involved.
Answer:
Yes. Amplification of nucleic acid through PCR even though micro-organisms are in low concentration.
Question 9.
Name any two scientists who were, credited for showing the role of penicillin as an antibiotic.
Answer:
Ernst Chain, Howard Florey and Alexander Hemming.
Question 10.
Name an immunosuppressive agent. From where is it obtained?
Answer:
Cyclosporin-A. It is obtained from Trichoderma polysporum.
Section – B
6 x 4 = 24
Note: Answer any six questions. Each answer may be limited to 20 lines.
Question 11.
Write any four physiological effects of cytokinins in plants.
Answer:
- Cytokinins induce cell division.
- They help to produce new leaves, chloroplasts in leaves, lateral shoot growth and adventitious shoot formation.
- They help to overcome the apical dominance.
- They delay the leaf senescence.
- They help in stomatal opening by increasing K ions in guard cells.
Question 12.
Explain different types of co-factors.
Answer:
Co-factors are of two types. They are:
a) Metal ion co-factors: Metallic cations get tightly attached to the apoenzyme are called metalloenzymes.
Ex: Cu2+ cytochrome oxidase.
b) Organic co-factors :
They are of two types.
1. Co-enzymes: They are the small organic molecules which are loosely associated with the Appoenzyme.
Ex: Thiamine pyrophosphate-Vitamin B1.
2. Prosthetic group: They are the organic co-factors which are tightly bound to the Apo-enzyme.
Ex: Haeme is the prosthetic group of enzyme Peroxidase.
![]()
Question 13.
Describe in brief photorespiration.
Answer:
Photorespiration is a light-dependent release of CO2 and uptake of O2 by the green tissue in C3 plants. It is also called oxidative photosynthetic carbon cycle or C2 photosynthesis. In this, RUBISCO (most abundant enzyme) binds with oxygen and hence CO2 fixation is decreased. Here RUBP binds with O2 to form one molecule of phosphoglycerate and one molecule of phosphoglycolate. In this process, no synthesis of ATP and NADPH.
Moreover, ATP is utilised and releases CO2. So this process is a wasteful process. It occurs with the involvement of three cell organelles namely chloroplast, peroxisome and mitochondria. In this process, nearly 25% of the CO2 is released out thus decreases photosynthesis. In C4 plants, photorespiration does not occur because they have mechanism that increase the concentration of CO2 at the enzyme active site.
Question 14.
Transpiration and photosynthesis – a compromise, explain.
Answer:
Transpiration is an indirect measurement of photosynthesis. When the stoma is open during transpiration loss, CO2 from outside may enter into cells, a limiting factor affecting the rate of photosynthesis.
In order to carry out photosynthesis, plants need to open its stomata to exchange of gases. It turns water and CO2 into Glucose. Opening the stomata though causes the loss of water, this evaporation causes water to be pulled up from the soil. An actively photosynthesising plant has an insatiable need for water. Photosynthesis is limited by available water which can be swiftly depleted by transpiration. C4 photosynthetic system is one of the structures for maximising the availability of CO2 and minimising water loss.
Question 15.
What is ICTV? How are viruses named?
Answer:
International Committee on Taxonomy of Viruses results the norms of classification and nomenclature of Viruses. The ICTV has only three hierarchial levels, the Family, Genus and Species. The family names end with the suffix ‘Viridae. While the Genus names end with Virus and the species names are with common English expressions. Viruses are named after the disease they cause. Ex: Polio Virus.
Question 16.
Mention the advantages of selecting pea plant for experiment by Mendel.
Answer:
Garden pea (Pisum sativum) is suitable for hybridization experiments due to the following advantages:
- It is an annual plant that has well-defined Characteristics.
- It can be grown and crossed easily.
- It has bisexual flowers containing both female and male parts. ,
- It can be self-fertilized conveniently.
- It has a short life cycle and produces large number of off springs.
Question 17.
What are the contributions of George Gamow, H.G. Khorana, Marshall Nirenberg in deciphering the genetic code?
Answer:
George Gamow: A physicist suggested that in order to code for all the 20 ammo acids, the code should be made up of three nucleotides. This was a very bold proposition because a permutation and combination of 43 (4 x 4 x 4) would generate 64 codons, generating many more codons than required.
H.G. Khorana: He developed chemical method in synthesising RNA molecules with defined combinations of bass (1-homopolymers such as UUU and co-polymers such as UUC, CCA.) Marshall Nirenberg: He proposed cell – free system for protein synthesis finally helped the code to be deciphered.
Question 18.
Give a brief account of pest-resistant plants.
Answer:
Several nematodes parasitize a wide variety of plants and mammals. A Nematode, Meloidegyne incognitia infects the roots of tobacco plants and cause a great reduction in yield. To prevent this infestation, process of RNA interference was adopted. Using Agrobacterium vectors, nematode-specific genes were introduced into the host plant.
The introduction of DNA was such that it produces both sense and anti-sense RNA ase in the host cells. These two RNAs forms a double-stranded RNA that initiated RNAi and thus silenced the specific mRNA of the nematode. The parasite could not survive in a transgenic host expressing specific interfering RNA. The transgenic plant therefore got itself protected from the parasite.
![]()
Section -C
2 x 8 = 16
Note: Answer any two questions. Each answer may be limited to 60 lines.
Question 19.
Explain the reactions of Kreb’s cycle.
Answer:
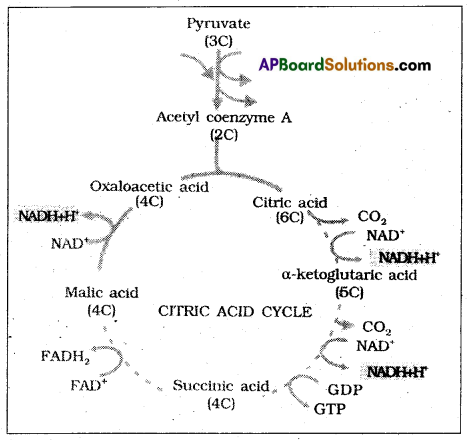
The acetyl CoA enters into the [initochondrial matrix] a cyclic pathway tricarboxylic acid cycle, more commonly called Krebs cycle after the scientist Hans Krebs who first elucidated it.
1. Condensation: In this acetyl, CoA condenses with oxaloacetic acid and water to yield citric acid in’ the presence of citrate synthetase and CoA is released.
![]()
2. Dehydration: Citric acid looses water molecules to yield cis aconitic acid in the presence of aconitase.
![]()
3. Hydration: A water molecule is added to cisaconic acid to yield isocitric acid in the presence of a conitase.
![]()
4. Oxidation I: Isocitric acid undergoes oxidation in the presence of dehydrogenase to yield succinic acid.

5. Decarboxylation: Oxalosuccinic acid undergoes decarboxylation in the presence of decarboxylase to form α-keto glutaric acid.

6. Oxidation if, decarboxylation: α – keto glutaric acid undergoes oxidation and decarboxylation in the presence of dehydrogenase and condenses with co.A to form succinyl Co. A.

7. Cleavage: Succinyl Co.A splits into succinic acid and Co.A in the presence of thiokinase to form succinic acid. The energy released is utilised to from ATP from ADP and PI.
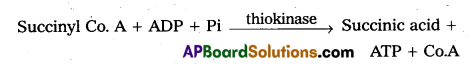
8. Oxidation – III: Succinic acid undergoes oxidation and forms Rimaric acid in the presence of succinic dehydrogenase.

9. Hydration: A water molecule is alcohol to Rimaric acid in the presence of Fumarase to form Malic acid.
![]()
10. Oxidation IV: Malic acid undergoes oxidation in the presence of malic dehydrogenase to form oxaloacetic acid.

In TCA cycle, for every 2 molecules of Acetyl Co.A undergoing oxidation, 2 ATP 8 NADPH+ H+, 2FADH2 molecules are formed.
Question 20.
Give a brief account of the tools of recombinant DNA technology.
Answer:
Key tools are:
1. Restriction enzymes: Two enzymes responsible for restricting the growth of Bacteriophage in Escherichia coli were isolated in the year 1963. One of these added methyl group to DNA and the other DNA. The latter was called restriction endonuclease. The first restriction endonuclease. Hind [1 which cut DNA molecule at a particular pair by recognising a specific sequence of six base pairs, called recognition sequence for Hind II. Today more than 900 restriction enzymes were isolated from over 200 strains of Bacteria each of which recognises a different recognition sequence.
E. CORI is a restriction enzyme in which, the first letter comes from the genus (escherichia) and the second two letters from the species of the prokaryotic cell (cou) the latter R’ is derived from the name of strain. Roman numbers indicate the order in which the enzymes were isolated from the strain of Bacteria. Restriction enzyme belong to a larger class of enzymes called nucleases. They are of two types:
- Exonucleases which remove nucleotides from the ends of the DNA.
- Endonucleases Which makes cuts at specific location within the DNA.
Most restriction enzymes cut the two stands of DNA double helx at different locations such a clevage is known as staggered cut. E.CORI recognises 5’ GAATI’3 sites on the DNA and cut it between G & A results in the formation of sticky ends or cohesive end pieces.
This stickyness of the ends facilitates the action of enzyme DNA ligase. Cloning vectors: The DNA used as a carrier for transferring a fragment of foreign DNA into a suitable host is called vector. Vectors used for multiplying the foreign DNA sequences are called cloning vectors. Commonly used cloning vectors are plasmids, bacterio-phages, cosmids, plasmids are extra chromosome circular DNA molecules present in almost all bacteria species. They are inheritable and carry few genes are easy to isolate and reintroduce into the bacterium (host).
Features required to facilitates cloning into a vector:
a) Origin of replication: (on) This is a sequence from where replication starts and any piece of DNA when linked to this sequence can be made to replicate within host cells. It is also responsible for controlling the copy number of the linked DNA.
b) Selectable marker: In addition to ‘on’ the vector requires a selectable maker, which help in identifying and eliminating non-transformants and selectively permitting the growth of the any transformants normally, the genes encoding resistance to Antibiotics such as Ampicillin, Chloramphenicol, tetra cycline or Kanamycin etc. are useful selectable makers for Ecoli.
c) Cloning sites: In order to link the alien DNA. the vector needs to have very few preferably single recognition sites for the restriction enzyme.
d) Molecular weight: The cloning vector should have low molecular weight.
e) Vectors for cloning genes in plants and animals: The tumour-inducing (Ti) plasmid of Agrobacterium tunifaciens has now been modified into a cloning vector such that it is no more pathogenic to plants. Similarly, retroviruses have also been disarmed and are now used to deliver desirable genes into animal cells.
![]()
Question 21.
Describe the tissue culture technique and what are the advantages of tissue culture.
Answer:
Key tools are:
1) Restriction enzymes: Two enzymes responsible for restricting the growth of Bacteriophage in Escherichia coli were isolated in the year 1963. One of these added methyl group to DNA and the other DNA. The latter was called restriction endonuclease. The first restriction endonuclease. Hind [1 which cut DNA molecule at a particular pair by recognising a specific sequence of six base pairs, called recognition sequence for Hind II. Today more than 900 restriction enzymes were isolated from over 200 strains of Bacteria each of which recognises a different recognition sequence.
E. CORI is a restriction enzyme in which, the first latter comes from the genus (escherichia) and the second two letters from the species of the prokaryotic cell (cou) the latter R’ is derived from the name of the strain. Roman numbers indicate the order in which the enzymes were isolated from the strain of Bacteria. Restriction enzyme belong to a larger class of enzymes called nucleases. They are of two types:
- Exonucleases which remove nucleotides from the ends of the DNA.
- Endonucleases Which makes cuts at specific location within the DNA.
Most restriction enzymes cut the two stands of DNA double helx at different locations such a clevage is known as staggered cut. E.CORI recognises 5’ GAATI’3 sites on the DNA and cut it between G & A results in the formation of sticky ends or cohesive end pieces.
This stickyness of the ends facilities the action of enzyme DNA ligase. Cloning vectors : The DNA used as a carrier for transferring a fragment of foreign DNA into a suitable host is called vector. Vectors used for multiplying the foreign DNA sequences are called cloning vectors. Commonly used cloning vectors are plasmids, bacterio-phages, cosmids, plasmids are extra chromosome circular DNA molecules present in almost all bacteria species. They are inheritable and carry few genes are easy to isolate and reintroduce into the bacterium (host).
Features required to facilitate cloning into a vector:
a) Origin of replication: (on) This is a sequence from where replication starts and any piece of DNA when linked to this sequence can be made to replicate within host cells. It is also responsible for controlling the copy number of the linked DNA.
b) Selectable marker: In addition to ‘on’ the vector requires a selectable maker, which help in identifying and eliminating non-transformants and selectively permitting the growth of the any transformants normally, the genes encoding resistance to Antibiotics such as Ampicillin, Chloramphenicol, tetra cycline or Kanamycin etc. are useful selectable makers for Ecoli.
c) Cloning sites: In order to link the alien DNA. the vector needs to have very few preferably single recognition sites for the restriction enzyme.
d) Molecular weight: The cloning vector should have low molecular weight.
e) Vectors for cloning genes in plants and animals: The tumour-inducing (Ti) plasmid of Agrobacterium tunifaciens has now been modified into a cloning vector such that it is no more pathogenic to plants. Similarly, retroviruses have also been disarmed and are now used to deliver desirable genes into animal cells.